Our papers
High-Yield Assembly of Plasmon-Coupled Nanodiamonds Using DNA Origami for Tuned Emission
The work of Niklas is published in Small Structures!
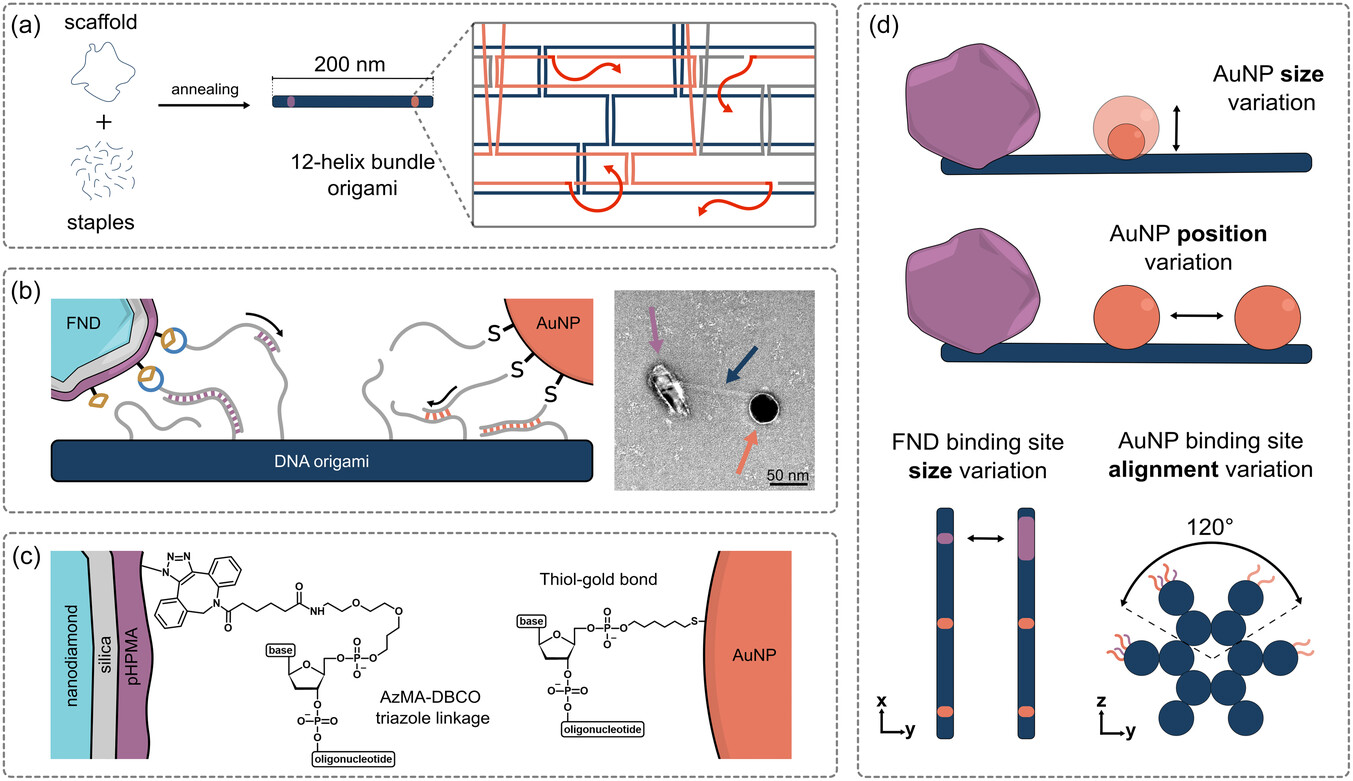
We present a DNA origami-based method for assembling nanodiamonds and gold nanoparticles
with unprecedented reliability and spatial control. Nanodiamonds hosting nitrogen-vacancy (NV)
centers are among the most promising solid-state quantum sensors: they can detect magnetic fields,
temperature, or electric signals at the nanoscale - even inside living cells. When coupled with
plasmonic nanoparticles like gold, their light emission can be tuned and enhanced, which can improve their performance.
Until now, building such hybrid structures with well-defined geometry and good reproducibility has been extremely
difficult. We use DNA origami as nanoscale scaffolds to solve this problem. We present a covalent
functionalization strategy to bind single-stranded DNA to nanodiamonds, which allows precise placement
of nanodiamonds and gold nanoparticles at controlled distances. This approach achieves assembly yields above
50% — a major step forward compared to previous random or low-yield methods.
By systematically varying the distance between gold and diamond nanoparticles, we observed a clear,
distance-dependent change in the photoluminescence lifetime of NV centers — direct evidence of efficient
coupling between the diamond's electronic states and the gold's localized plasmons.
Beyond advancing the study of NV-plasmon interactions, this method opens new pathways
toward scalable architectures for quantum photonics, nanoscale sensing, and energy transfer studies.
This method can be particularly useful for validating any sensors based on nanodiamonds.
Reach out to us if you are interested!
This work was possible thanks to the fantastic collaboration with Petr Cígler, Jakub Čopák and Marek Kindermann who developed the chemistry to functionalize nanodiamonds with DNA.
Optimized molecule detection in localization microscopy with selected false positive probability
Mirek's paper about detecting molecules in microscopy images out in Nature Communications!
We present a method to detect molecules in microscopy images using probabilistic thresholding.
With only one parameter to set conditions and describe error levels, it simplifies the process
for users and enables reproducibility.
Inspired by radars and astronomy, we used signal detection theory to design an optimal filter for microscopy and
implement probabilistic thresholding that allows to control the level of false positive detections.
Why is this important? Detection of molecules is an essential first step in Single Molecule Localization Microscopy.
Yet, it currently lacks quantification of detection errors resulting in artifacts and affecting further analysis.
Furthermore, it is unduly complex.
How did we solve it? By combining two steps. First, we derived a theoretically optimal filter for Poisson
noise prevalent in microscopy, Poisson Matched Filter, and used it as a performance benchmark for other
filters used in SMLM, setting Matched Filter as a desired option. Second, we implemented probabilistic
thresholding, where the level of false positive probability sets the detection threshold and minimizes
false negative detections. To enhance robustness against varying background, we set the threshold adaptively.
We will be happy to collaborate with developers of SMLM software on implementing the method and bringing it to the users! Please,
reach out to us!
Please check our further publications at